| Changes from version 2 to version 3.1 (Sep 2017) |
| An RNA-seq extension strategy is used to improve the annotation of 3' ends of genes. |
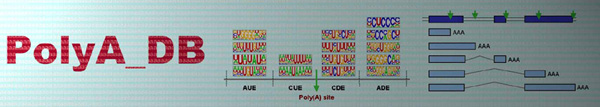
| PASs are identified using 3'READS sequencing data. |
| Over 100 samples from various cell lines and tissues of in human, mouse, rat and chicken are used. |
| Conservation information is assigned to each PAS based on syntenic regions and reciprocal best matches. |